Network Analysis
DA 101, Dr. Ladd
Week 12
What are Networks?
Networks are made up of…
- Entities (entity = node/vertex/actor)
- Relationships (relationship = edge/link/tie)
- We’ll use “nodes” and “edges”
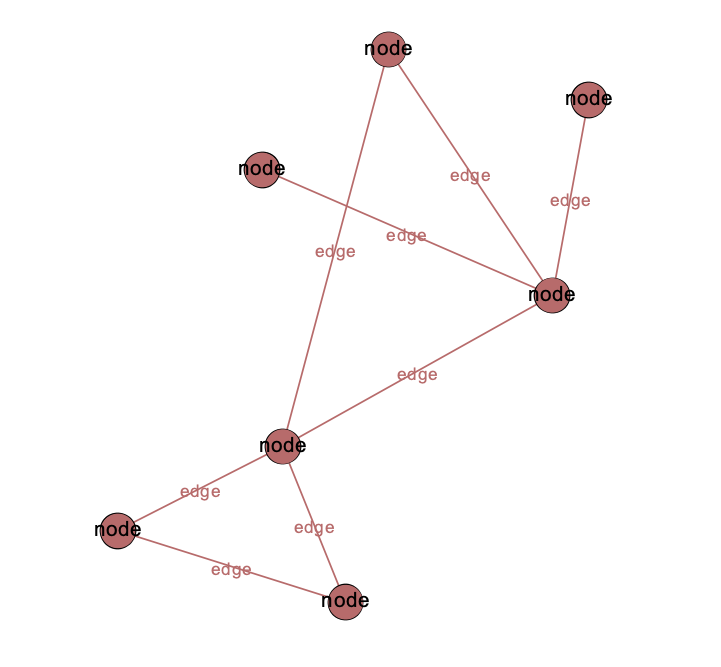
Nodes and Edges have Attributes
Node Attributes
- numerical (size)
- categorical (color)
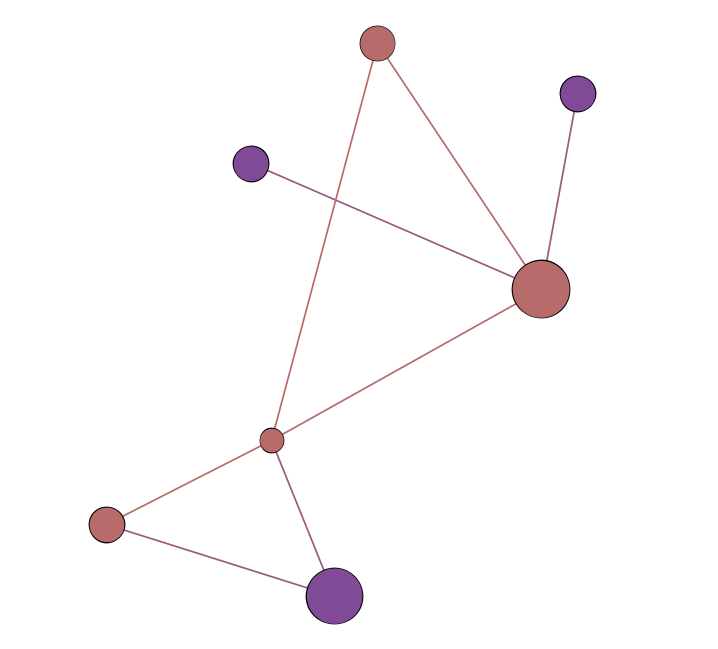
Edge Attributes
Directed and Undirected Edges
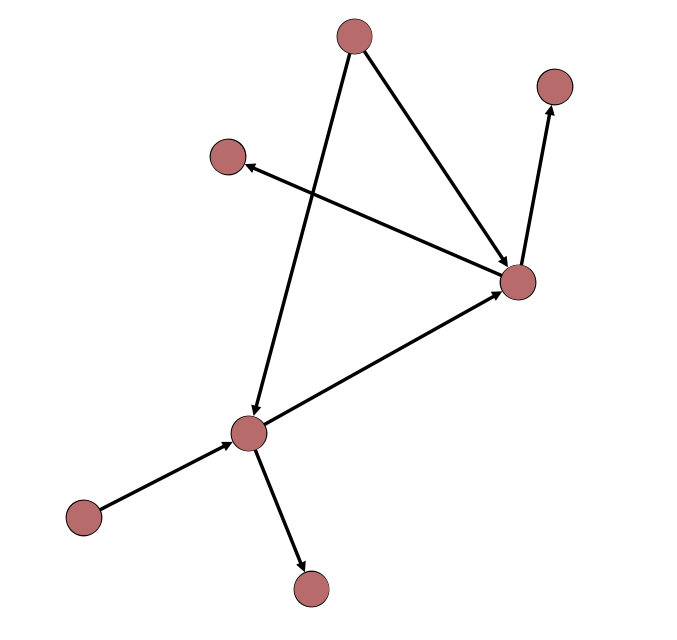
Weighted and Unweighted Edges
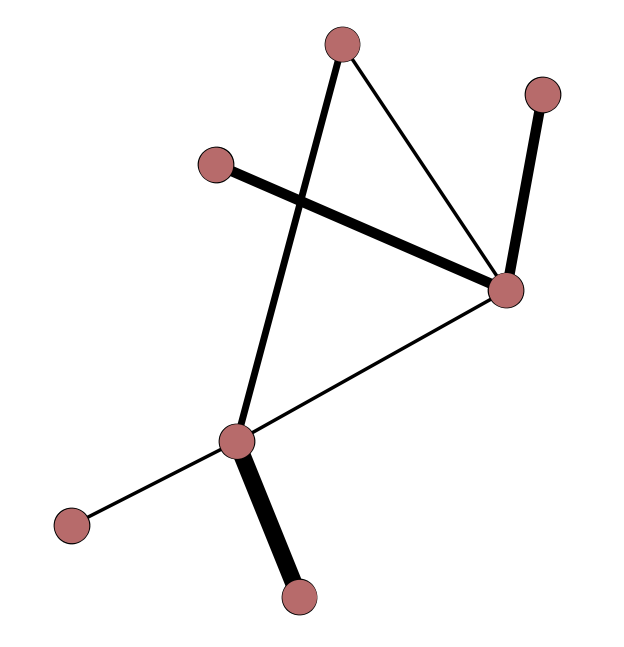
Edge Types
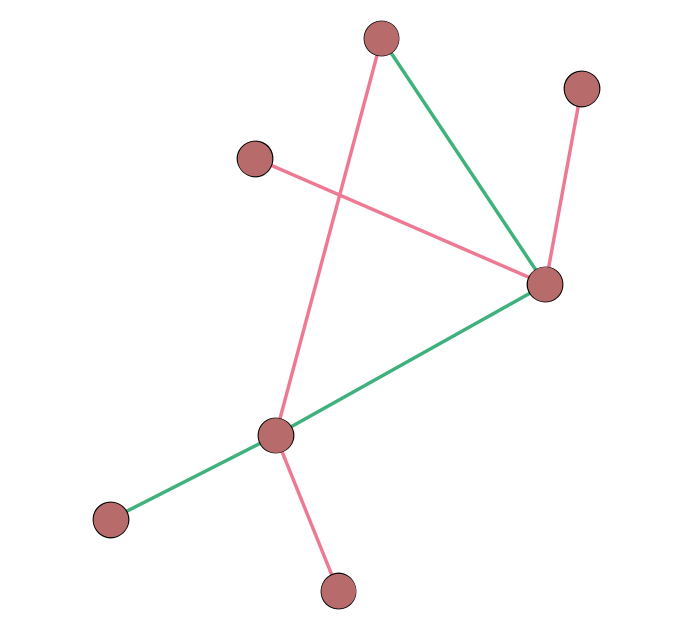
Multiple Edges “in a row” Make a Path
Path & Diameter
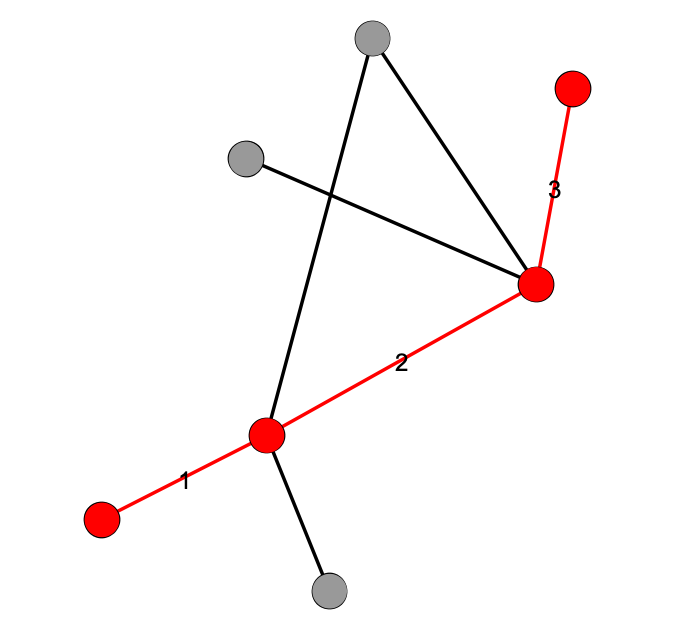
(& Average Shortest Path Length)
Some special kinds of nodes
Isolates
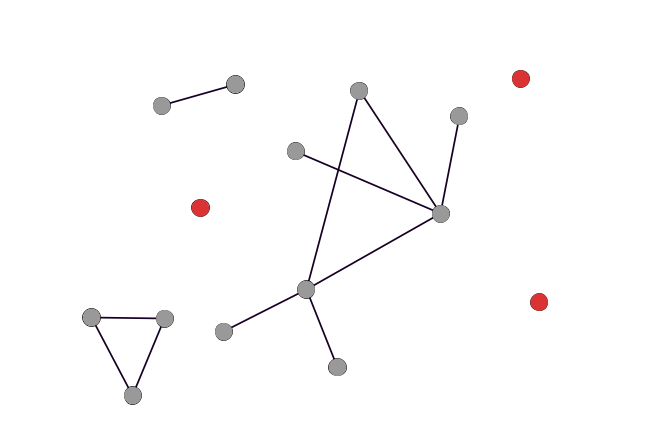
Hubs
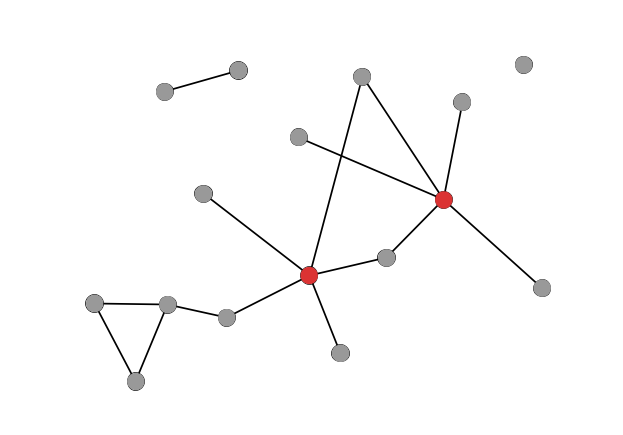
Bridges

Measuring a node’s “importance” with centrality
Degree
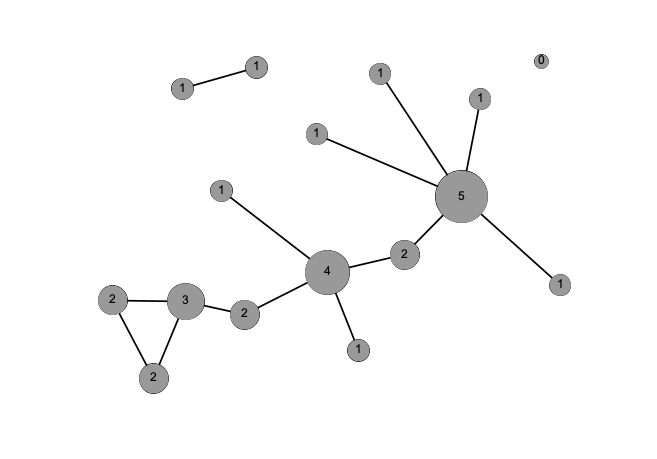
Strength
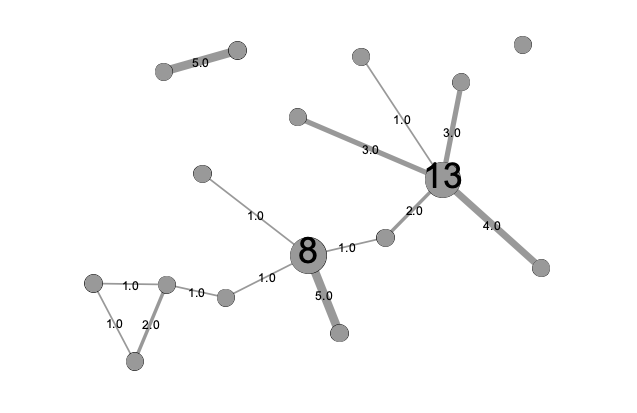
Betweenness
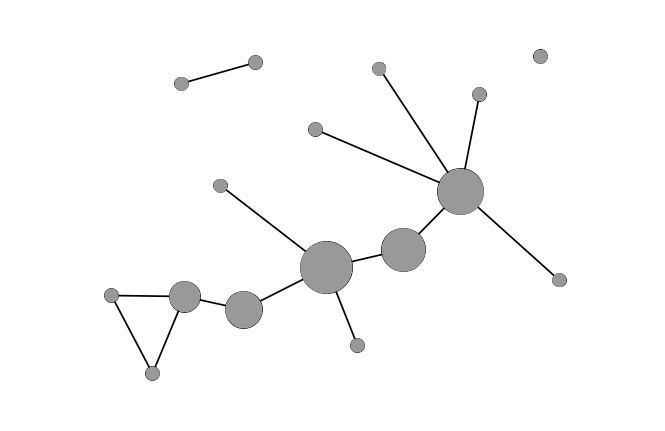
Different kinds of entities or nodes
Unipartite/unimodal
Bipartite/bimodal
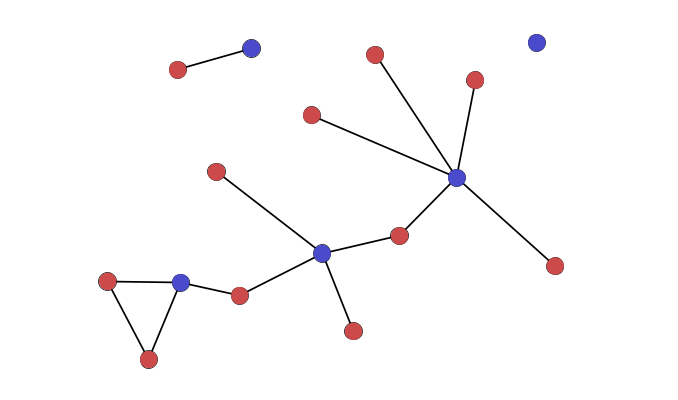
Bipartite (cont.)
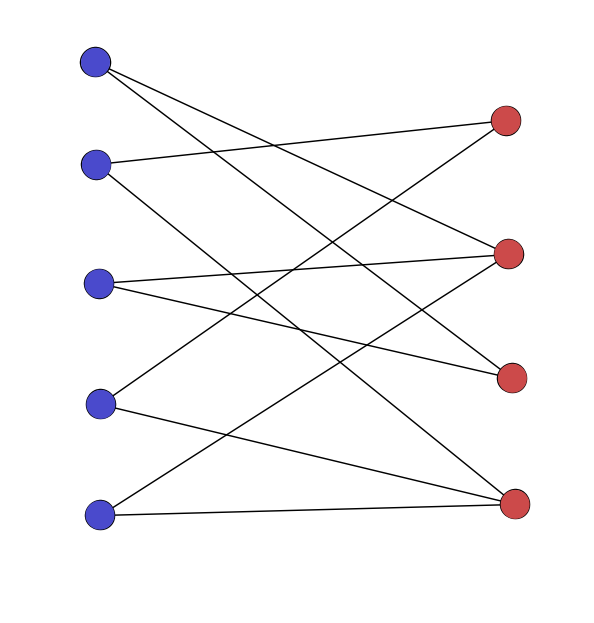
Multipartite/k-partite/multimodal
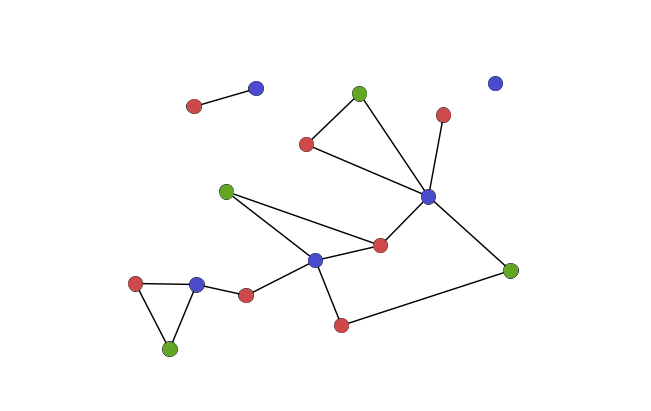
Groups of nodes within a network
Connected components
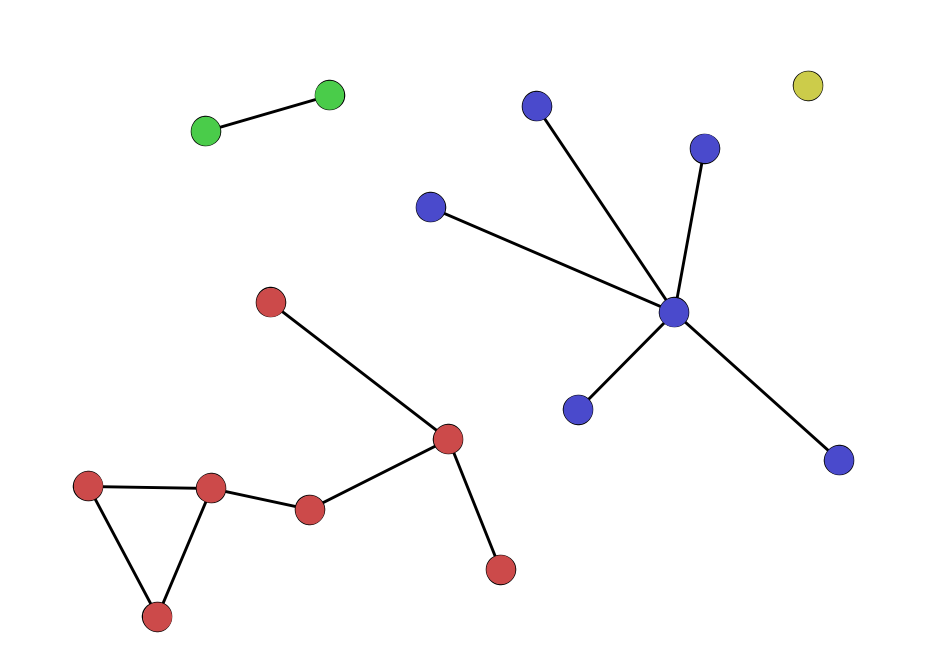
Cliques and clustering
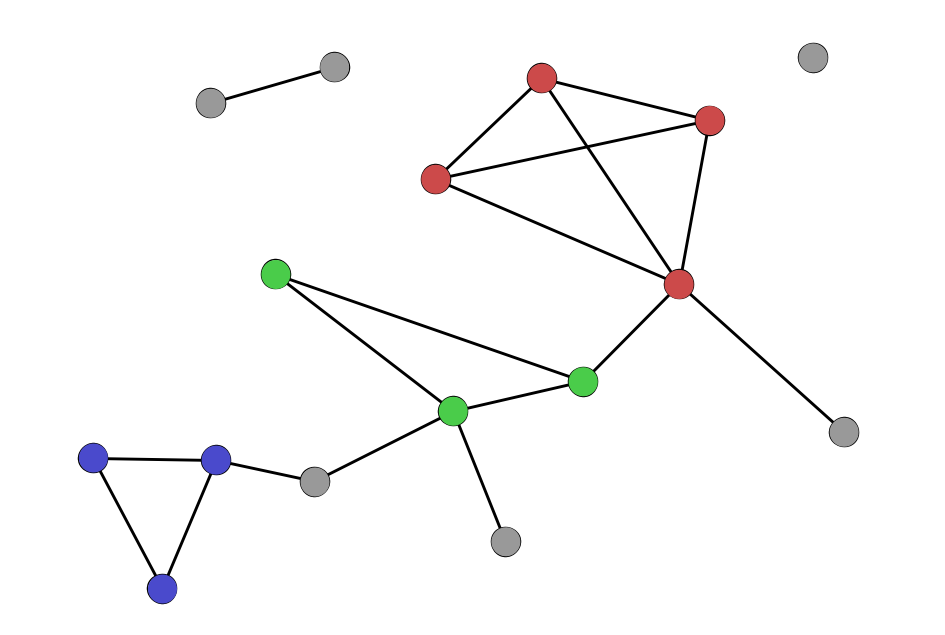
Clustering Coefficient
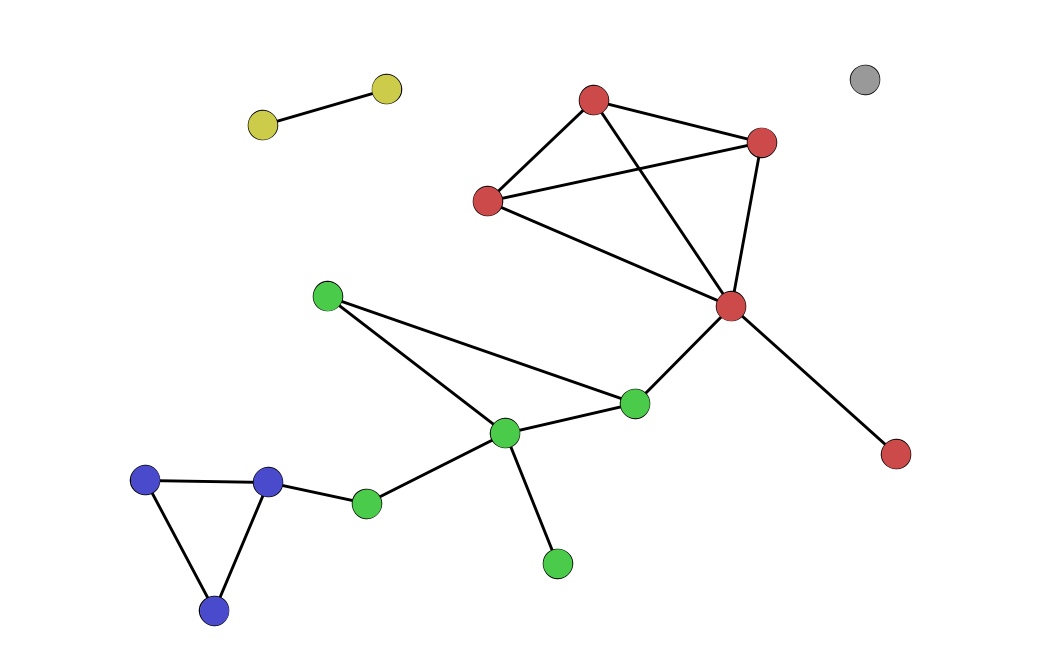
Communities and community detection
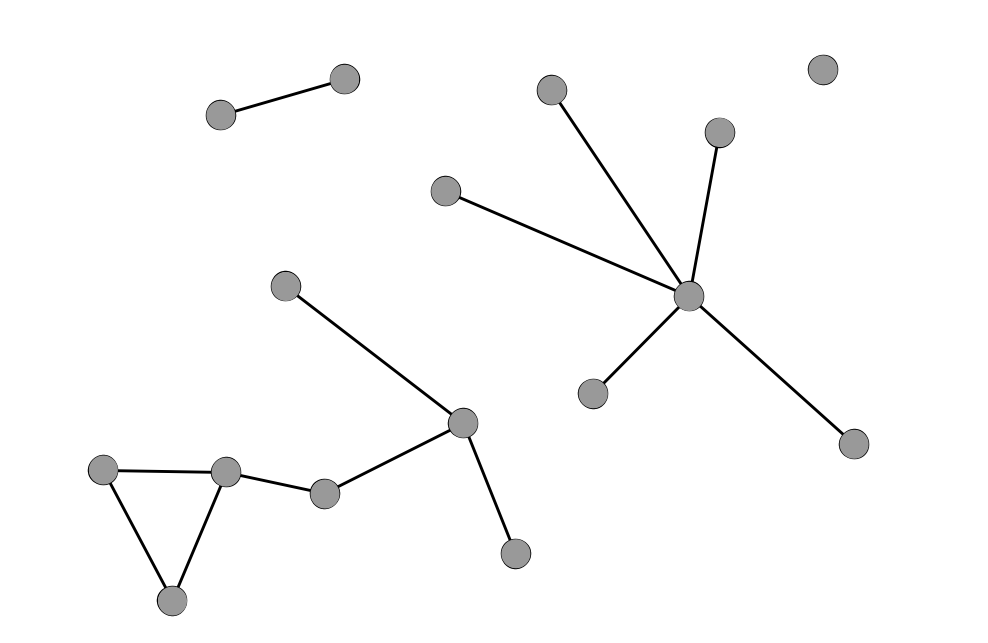
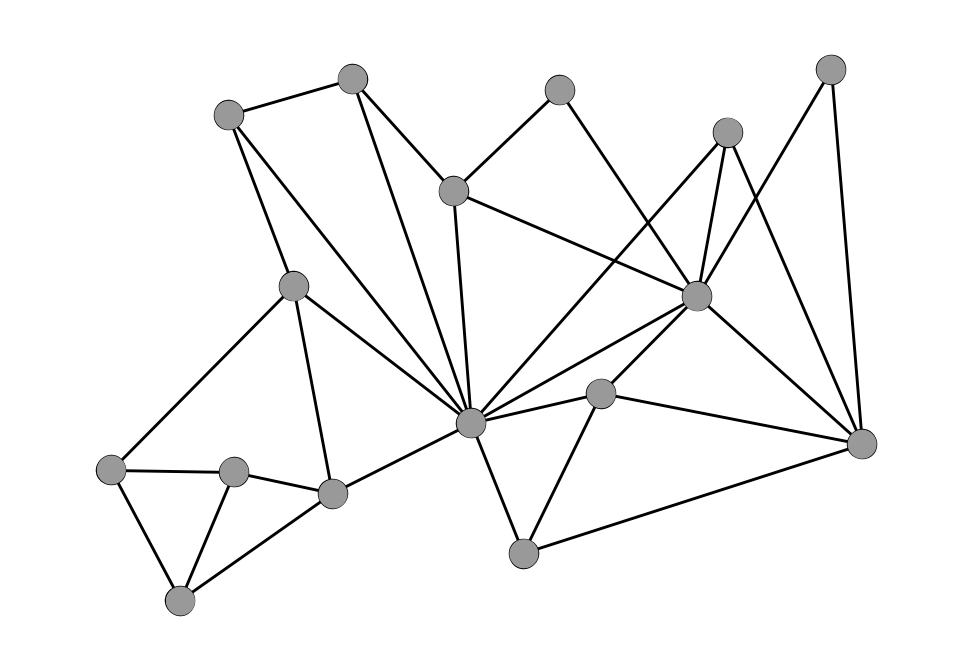
Density
A Sparse Network
A Dense Network
There are many ways to visualize a network
Adjacency Matrix
Adjacency List
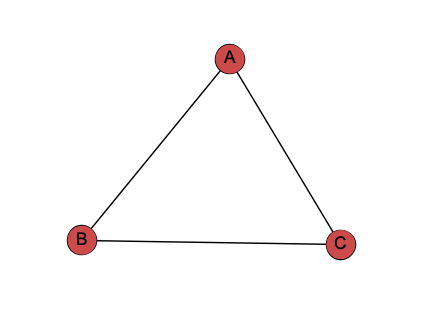
- A adjacent to B,C
- B adjacent to A,C
- C adjacent to A,B
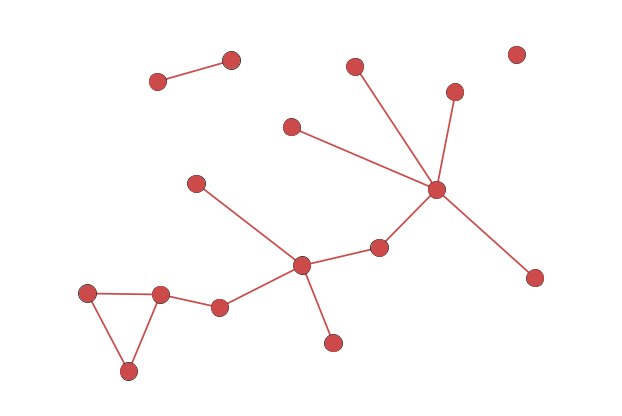
Node-Link Diagram

Other Important Concepts
Triadic Closure

Assortative mixing/Homophily
Preferential Attachment
Weak Ties
Small World Network

- low average path length
- low clustering coefficients
- degree distribution follows power law (a few large hubs)
- low diameter (usually around “six degrees”)
Working with Networks in R
Let’s start with an example.

We’ll need two new libraries.
We can download the data and load it into a “network object” with iGraph.
First, download got-edges.csv. What does this data look like?
Let’s make two versions of a node-link diagram.
We can use iGraph to calculate different metrics.
# Calculate degree and add to network data
V(G)$degree <- degree(G)
# Calculate betweenness centrality and add to network data
V(G)$betweenness <- betweenness(G, normalized=TRUE)
# Calculate the ratio of betweenness to degree
# (i.e. This is the node that functions most as a "bridge")
V(G)$betweenness_over_degree <- betweenness(G)/degree(G)You can convert node/vertex data to a dataframe.
What happens if you try:
With a dataframe, you can easily see degree and betweenness distributions.
How would you make a graph of the betweenness distribution?
Finally, let’s make a better node-link diagram.
# Convert data to networkD3-friendly format & calculate modularity clusters
G_d3 <- igraph_to_networkD3(G, membership(cluster_fast_greedy(G)))
# Add degree to node data and weight to link data
G_d3$nodes$degree <- V(G)$degree
G_d3$links$weight <- E(G)$weight
# Create force network with nodes sized by degree and colored by modularity
forceNetwork(Links=G_d3$links, Nodes = G_d3$nodes,
Source = 'source', Target = 'target',
Value= 'weight', NodeID = 'name',
Group = 'group', Nodesize='degree',
zoom = TRUE, opacity = 1)